Data manipulation basics
Overview
- The
tidyverse- R Packages
- Importing data
- The
dplyrpackagefilter()mutate()ifelse()- pipes
|> summarize()group_by()
- The
tidydata format
The tidyverse
Packages

Packages
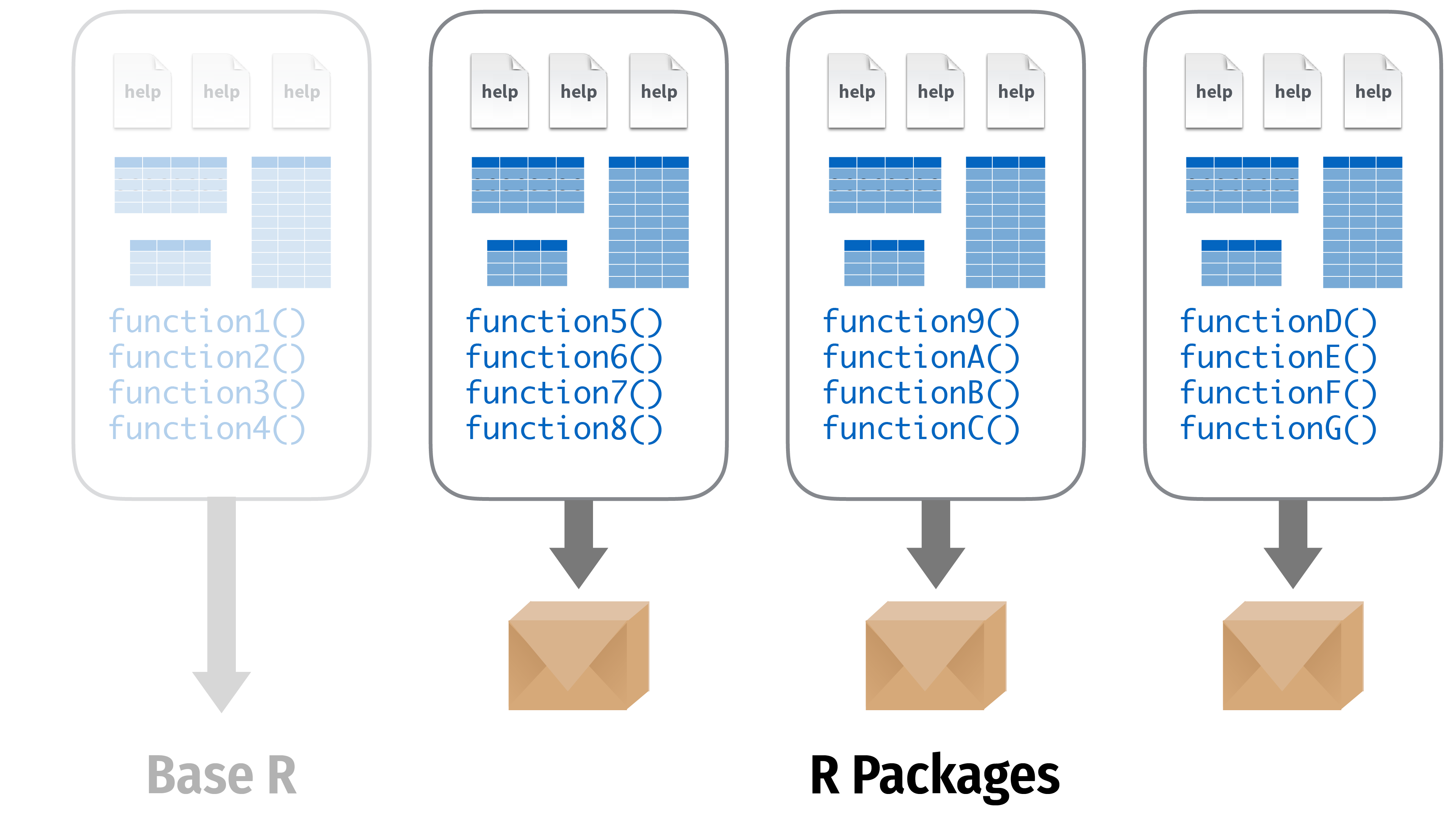
Packages
- So far we only used functions that are directly available in R
- But there are tons of user-created functions out there that can make your life so much easier
- These functions are shared in what we call packages
- Packages are bundles of functions that R users put at the disposal of other R users
- Packages are centralized on the Comprehensive R Archive Network (CRAN)
- To download and install a CRAN package you can simply type `install.packages()
Using packages
The tidyverse
“The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.”
… the tidyverse makes data science faster, easier and more fun…
The tidyverse

The tidyverse
The tidyverse package is a shortcut for installing and loading all the key tidyverse packages
The tidyverse
Installs all of these:
install.packages("ggplot2")
install.packages("dplyr")
install.packages("tidyr")
install.packages("readr")
install.packages("purrr")
install.packages("tibble")
install.packages("stringr")
install.packages("forcats")
install.packages("lubridate")
install.packages("hms")
install.packages("DBI")
install.packages("haven")
install.packages("httr")
install.packages("jsonlite")
install.packages("readxl")
install.packages("rvest")
install.packages("xml2")
install.packages("modelr")
install.packages("broom")Importing data

|
Work with plain text data |
my_data <- read_csv(“file.csv”)
|

|
Work with Excel files |
my_data <- read_excel(“file.xlsx”)
|

|
Work with Stata, SPSS, and SAS data |
my_data <- read_stata(“file.dta”)
|
Data from R-Packages
Some data sets can be downloaded as packages in R. For example, the gapminder data set.
Install the package
Then load the data
The dplyr package
tidyverse
dplyr


dplyr: verbs for manipulating data
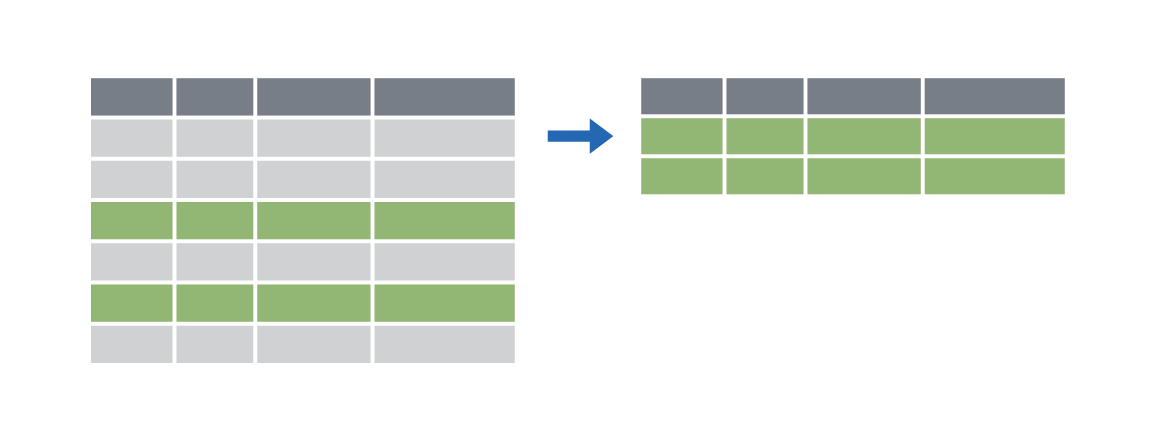
Extract rows with filter()
|
|
Extract columns with select()
|
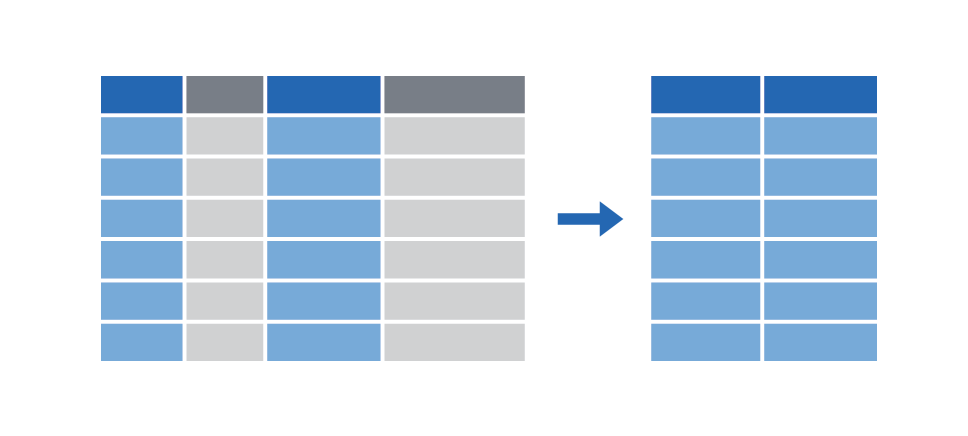
|
Arrange/sort rows with arrange()
|
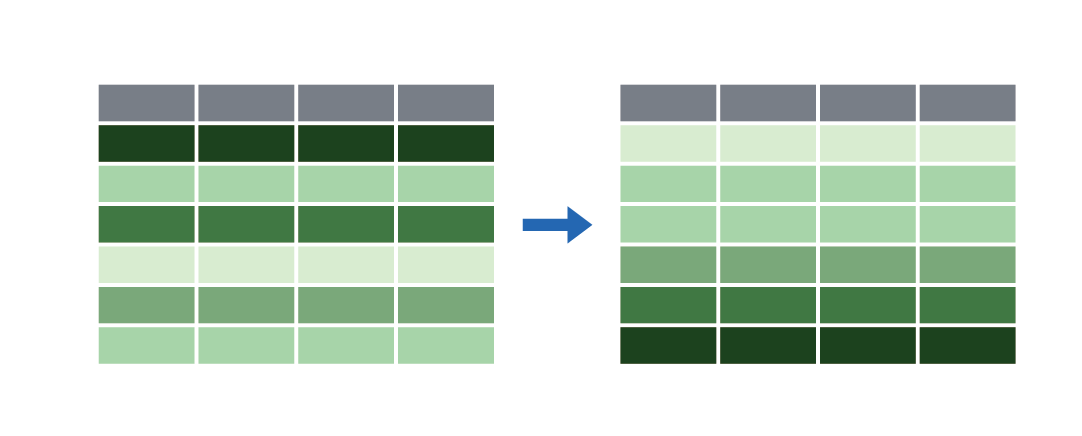
|
Make new columns with mutate()
|
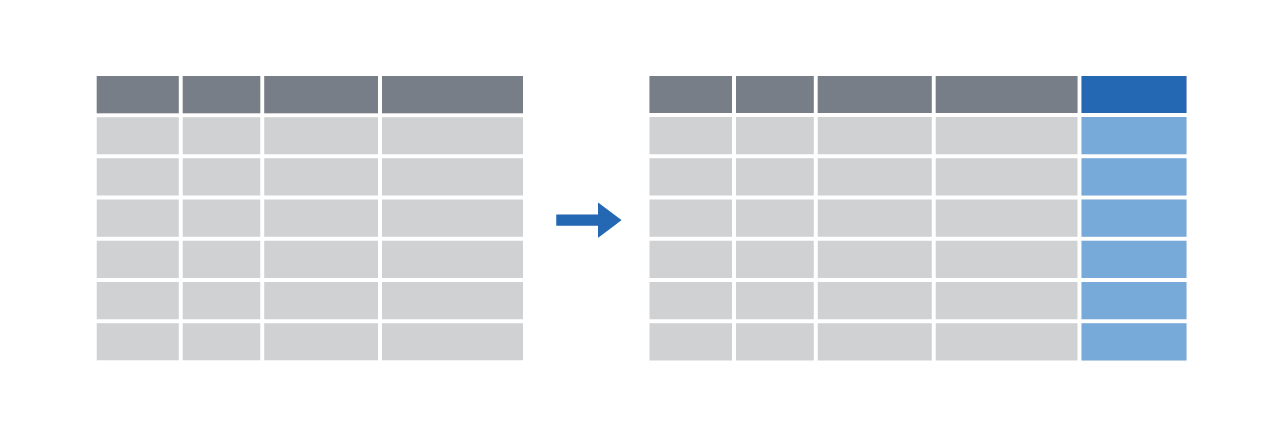
|
Make group summaries with group_by() |> summarize()
|
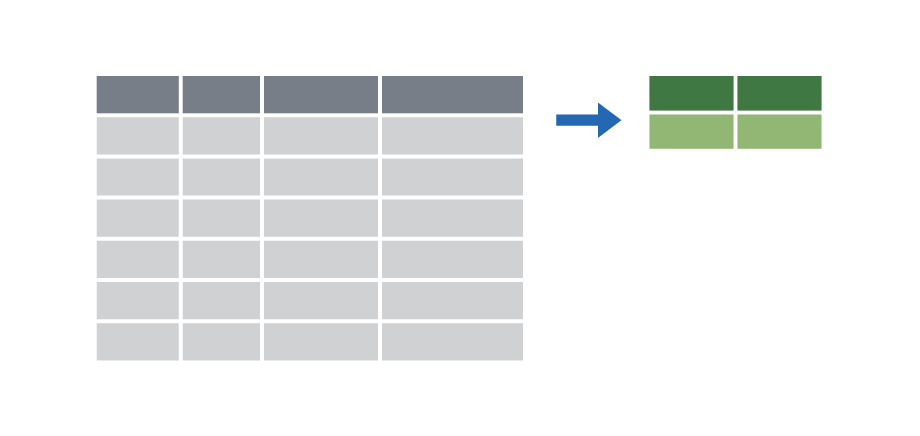
|
filter()
Extract rows that meet some sort of test
Logical tests
| Test | Meaning | Test | Meaning |
|---|---|---|---|
x < y |
Less than | x %in% y |
In (group membership) |
x > y |
Greater than | is.na(x) |
Is missing |
== |
Equal to | !is.na(x) |
Is not missing |
x <= y |
Less than or equal to | ||
x >= y |
Greater than or equal to | ||
x != y |
Not equal to |
Your turn #1: Filtering
Use filter() and logical tests to show…
- The data for Canada
- All data for countries in Oceania
- Rows where the life expectancy is greater than 82
04:00
Your turn #1: Filtering
Use filter() and logical tests to show…
Common Mistakes
Using = instead of ==
Forgetting quotes ("")
filter() with multiple conditions
Extract rows that meet every test
# A tibble: 2 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Denmark Europe 2002 77.2 5374693 32167.
2 Denmark Europe 2007 78.3 5468120 35278.Boolean operators
| Operator | Meaning |
|---|---|
a & b |
and |
a | b |
or |
!a |
not |
Boolean operators
The default is “and”
These do the same thing:
Your turn #2: Filtering
Use filter() and Boolean logical tests to show…
- Canada before 1970
- Countries where life expectancy in 2007 is below 50
- Countries where life expectancy in 2007 is below 50 and are not in Africa
04:00
Your turn #2: Filtering
Use filter() and Boolean logical tests to show…
- Canada before 1970
- Countries where life expectancy in 2007 is below 50
- Countries where life expectancy in 2007 is below 50 and are not in Africa
Common Mistakes
Collapsing multiple tests into one
Using multiple tests instead of %in%
Common Syntax
Every dplyr verb function follows the same pattern
mutate()
Create new columns
mutate()
Create new columns
We can also create multiple new columns at once
| country | year | … | gdp | pop_mil |
|---|---|---|---|---|
| Afghanistan | 1952 | … | 6567086330 | 8 |
| Afghanistan | 1957 | … | 7585448670 | 9 |
| Afghanistan | 1962 | … | 8758855797 | 10 |
| Afghanistan | 1967 | … | 9648014150 | 12 |
| Afghanistan | 1972 | … | 9678553274 | 13 |
| Afghanistan | 1977 | … | 11697659231 | 15 |
ifelse()
Do conditional tests within mutate()
ifelse()
The new variable can take any sort of class
Your turn #3: Mutating
Use mutate() to…
- Add an
africacolumn that is TRUE if the country is on the African continent - Add a column for logged GDP per capita (hint: use
log()) - Add an
africa_asiacolumn that says “Africa or Asia” if the country is in Africa or Asia, and “Not Africa or Asia” if it’s not
05:00
Your turn #3: Mutating
Use mutate() to…
- Add an
africacolumn that is TRUE if the country is on the African continent
- Add a column for logged GDP per capita (hint: use
log())
What if you have multiple verbs?
Solution 1: Intermediate variables
Solution 2: Nested functions
|>
Why using pipes?
… 🤯 not easy to read
|> vs %>%
There are actually multiple pipes!
%>%was invented first, but requires a package to use|>is part of base RThey’re interchangeable 99% of the time (Just be consistent)
You do not have to type the pipe by hand every time
You can use the shortcut cmd + shift + m in R Studio.
summarize()
Compute a table of summaries
- Take a data frame
| country | continent | year | lifeExp |
|---|---|---|---|
| Afghanistan | Asia | 1952 | 28.801 |
| Afghanistan | Asia | 1957 | 30.332 |
| Afghanistan | Asia | 1962 | 31.997 |
| Afghanistan | Asia | 1967 | 34.02 |
| … | … | … | … |
Your turn #4: Summarizing
Use summarize() to calculate…
- The first (minimum) year in the dataset
- The last (maximum) year in the dataset
- The number of rows in the dataset (use the cheatsheet)
- The number of distinct countries in the dataset (use the cheatsheet)
05:00
Your turn #4: Summarizing
One Solution for all:
Your turn #5: Summarizing
Use filter() and summarize() to calculate…
- the number of unique countries and
- the median life expectancy
on the African continent in 2007.
05:00
Your turn #5: Summarizing
Use filter() and summarize() to calculate…
- the number of unique countries and
- the median life expectancy
on the African continent in 2007.
group_by()
Put rows into groups based on values in a column
Nothing happens by itself!
Powerful when combined with
summarize()
group_by()
| country | continent | year | lifeExp |
|---|---|---|---|
| Afghanistan | Asia | 1952 | 28.801 |
| Afghanistan | Asia | 1957 | 30.332 |
| Afghanistan | Asia | 1962 | 31.997 |
| Afghanistan | Asia | 1967 | 34.02 |
| … | … | … | … |
A simple summary
# A tibble: 1 × 1
n_countries
<int>
1 142Your turn #6: Grouping and summarizing
- Find the minimum, maximum, and median life expectancy for each continent
- Find the minimum, maximum, and median life expectancy for each continent in 2007 only
05:00
Your turn #6: Grouping and summarizing
- Find the minimum, maximum, and median life expectancy for each continent
- Find the minimum, maximum, and median life expectancy for each continent in 2007 only
dplyr: verbs for manipulating data
Extract rows with filter()
|

|
Extract columns with select()
|

|
Arrange/sort rows with arrange()
|

|
Make new columns with mutate()
|

|
Make group summaries with group_by() |> summarize()
|

|
Tidy data
Tidy data
You can represent the same underlying data in multiple ways.
# A tibble: 6 × 4
country year cases population
<chr> <dbl> <dbl> <dbl>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583# A tibble: 12 × 4
country year type count
<chr> <dbl> <chr> <dbl>
1 Afghanistan 1999 cases 745
2 Afghanistan 1999 population 19987071
3 Afghanistan 2000 cases 2666
4 Afghanistan 2000 population 20595360
5 Brazil 1999 cases 37737
6 Brazil 1999 population 172006362
7 Brazil 2000 cases 80488
8 Brazil 2000 population 174504898
9 China 1999 cases 212258
10 China 1999 population 1272915272
11 China 2000 cases 213766
12 China 2000 population 1280428583# A tibble: 6 × 3
country year rate
<chr> <dbl> <chr>
1 Afghanistan 1999 745/19987071
2 Afghanistan 2000 2666/20595360
3 Brazil 1999 37737/172006362
4 Brazil 2000 80488/174504898
5 China 1999 212258/1272915272
6 China 2000 213766/1280428583Tidy data
Tidy data has the following properties:
# A tibble: 6 × 4
country year cases population
<chr> <dbl> <dbl> <dbl>
1 Afghanistan 1999 745 19987071
2 Afghanistan 2000 2666 20595360
3 Brazil 1999 37737 172006362
4 Brazil 2000 80488 174504898
5 China 1999 212258 1272915272
6 China 2000 213766 1280428583
- Variables are columns
- Observations are rows
- Values are cells
Why ensure that your data is tidy?
There are two main advantages:
- There’s a general advantage to picking one consistent way of storing data. If you have a consistent data structure, it’s easier to learn the tools that work with it because they have an underlying uniformity.
- There’s a specific advantage to placing variables in columns because it allows R’s vectorized nature to shine. As you learned in ?@sec-mutate and ?@sec-summarize, most built-in R functions work with vectors of values. That makes transforming tidy data feel particularly natural.
dplyr, ggplot2, and all the other packages in the tidyverse are designed to work with tidy data.
Will I ever encounter a dataset that isn’t tidy?
Yes, unfortunately, most real data is untidy.
There are two main reasons:
Data is often organized to facilitate some goal other than analysis. For example, it’s common for data to be structured to make data entry, not analysis, easy.
Most people aren’t familiar with the principles of tidy data, and it’s hard to derive them yourself unless you spend a lot of time working with data.
Pivoting data
tidyr provides two main functions to “pivot” data in a tidy format:
pivot_longer()
and
pivot_wider()
Here, we’ll only discuss pivot_longer() because it’s the most common case.
pivot_longer()
- Suppose we have three patients with
ids A, B, and C, and we take two blood pressure measurements on each patient. - We’ll create the data with
tribble(), a handy function for constructing small tibbles by hand:
pivot_longer()
- We want our new dataset to have three variables:
id(already exists),measurement(the column names), andvalue(the cell values) - To achieve this, we need to pivot
dflonger
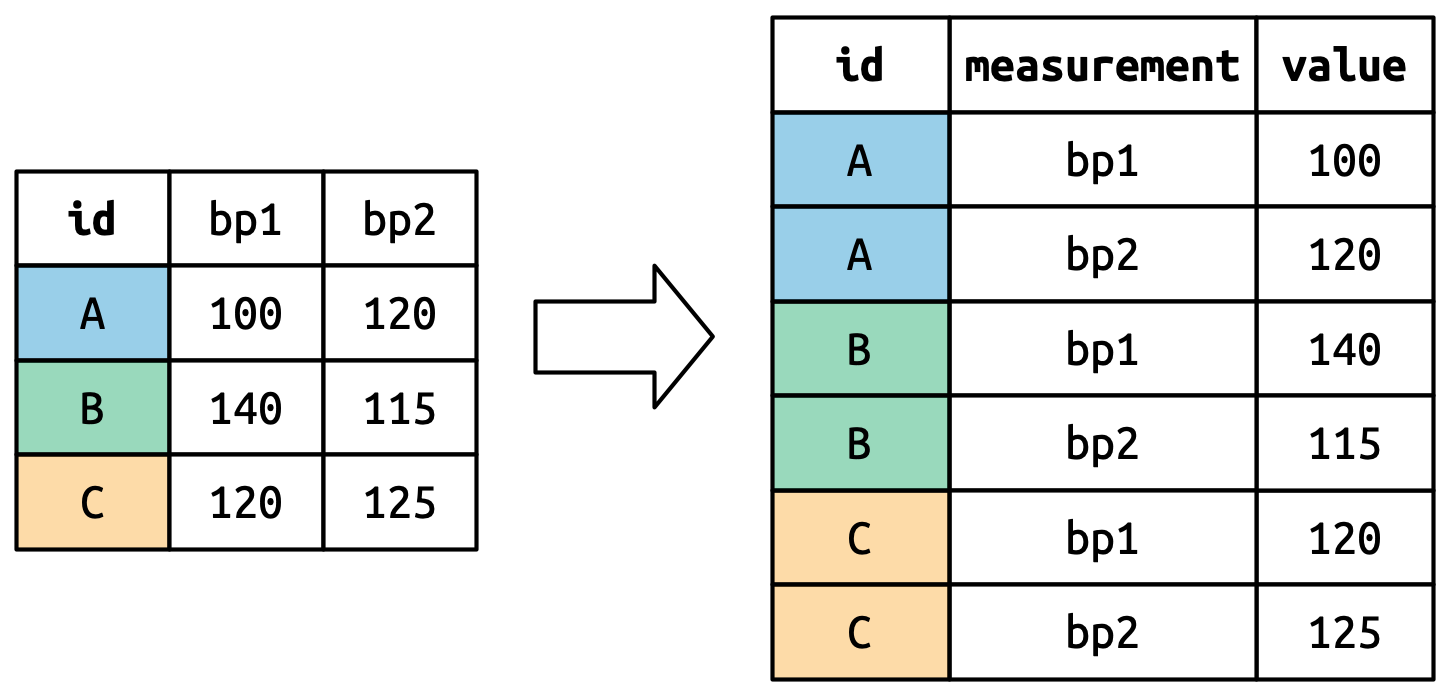
pivot_longer()
- The values in a column that was already a variable in the original dataset (
id) need to be repeated, once for each column that is pivoted.

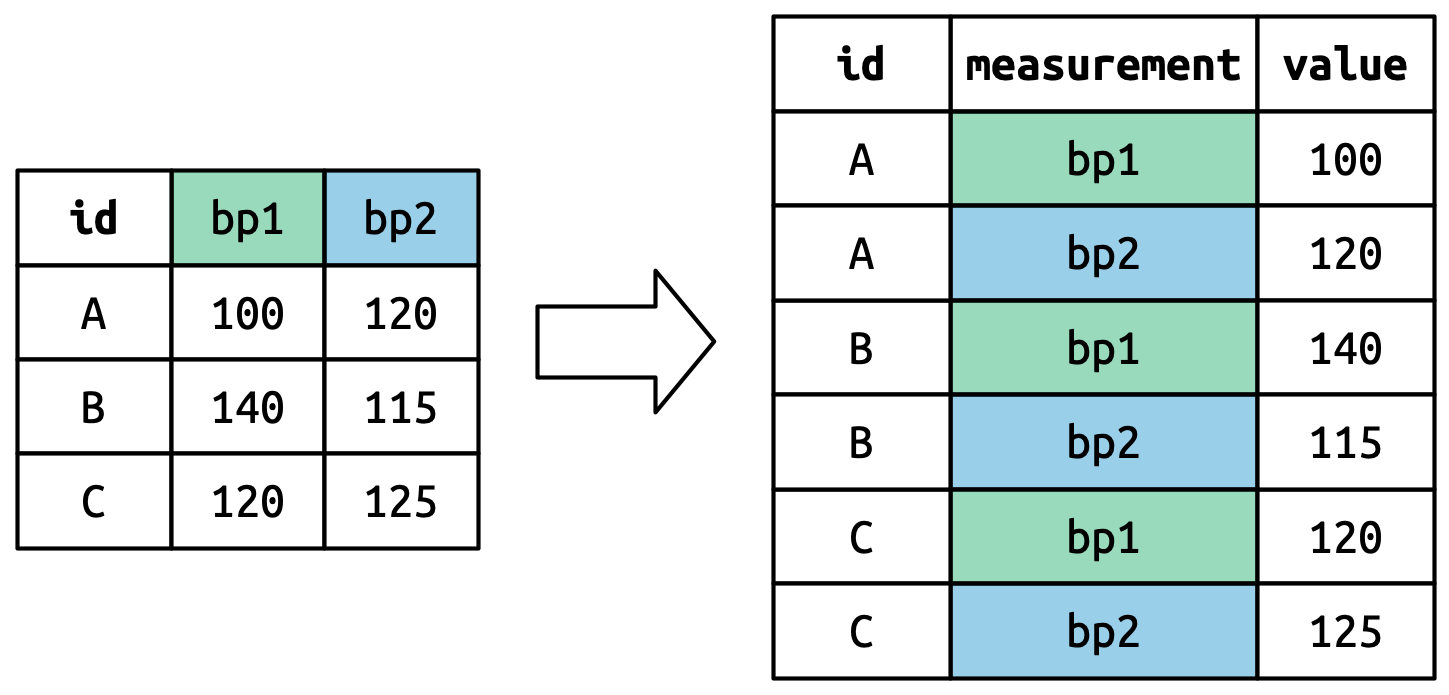
pivot_longer()
- The column names become values of the new variable
measurement
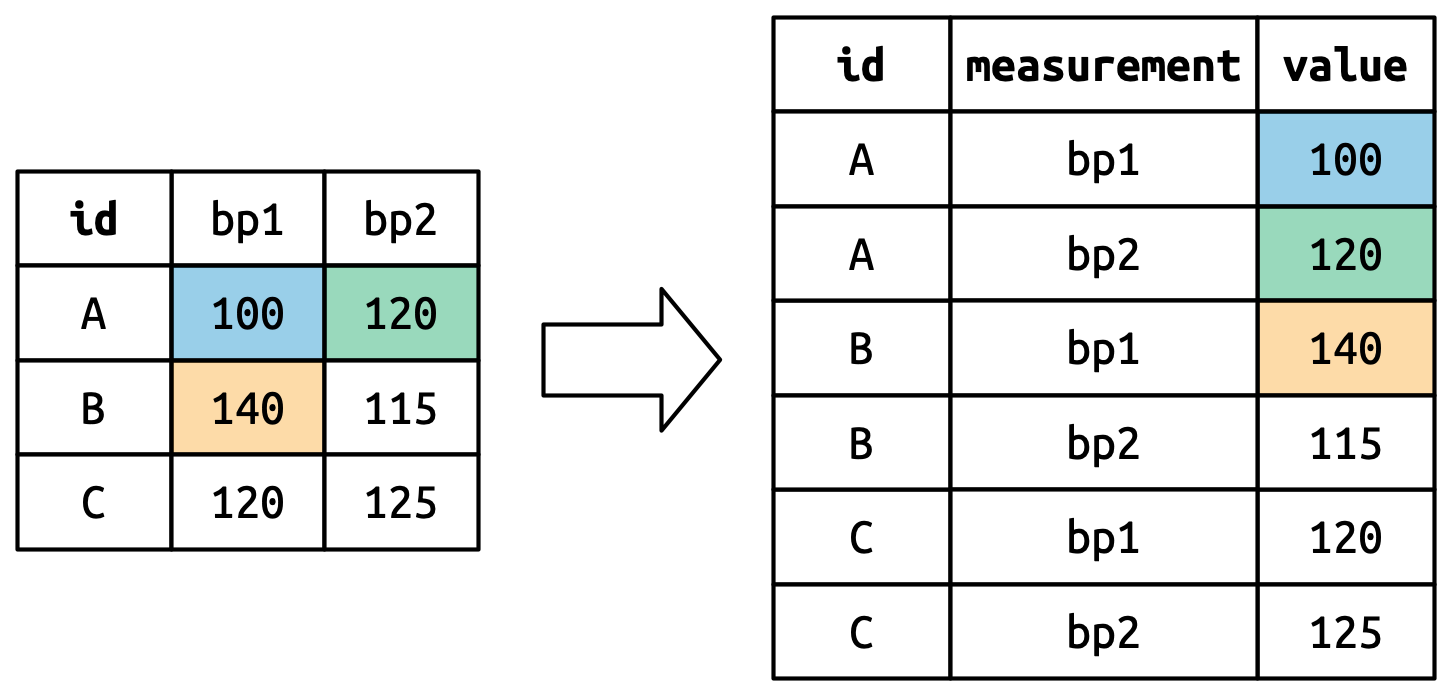
pivot_longer()
- The cell values become values of the new variable
value
pivot_longer()
# A tibble: 3 × 3
id bp1 bp2
<chr> <dbl> <dbl>
1 A 100 120
2 B 140 115
3 C 120 125There are three key arguments:
colsspecifies which columns need to be pivoted, i.e. which columns aren’t variables. This argument uses the same syntax asselect()names_tonames the variable in which column names should be storedvalues_tonames the variable in which cell values should be stored
Your turn #7: Pivoting
The billboard dataset which comes with the tidyverse package records the billboard rank of songs in the year 2000.
# A tibble: 6 × 79
artist track date.entered wk1 wk2 wk3 wk4 wk5 wk6 wk7 wk8
<chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2 Pac Baby… 2000-02-26 87 82 72 77 87 94 99 NA
2 2Ge+her The … 2000-09-02 91 87 92 NA NA NA NA NA
3 3 Doors Do… Kryp… 2000-04-08 81 70 68 67 66 57 54 53
4 3 Doors Do… Loser 2000-10-21 76 76 72 69 67 65 55 59
5 504 Boyz Wobb… 2000-04-15 57 34 25 17 17 31 36 49
6 98^0 Give… 2000-08-19 51 39 34 26 26 19 2 2
# ℹ 68 more variables: wk9 <dbl>, wk10 <dbl>, wk11 <dbl>, wk12 <dbl>,
# wk13 <dbl>, wk14 <dbl>, wk15 <dbl>, wk16 <dbl>, wk17 <dbl>, wk18 <dbl>,
# wk19 <dbl>, wk20 <dbl>, wk21 <dbl>, wk22 <dbl>, wk23 <dbl>, wk24 <dbl>,
# wk25 <dbl>, wk26 <dbl>, wk27 <dbl>, wk28 <dbl>, wk29 <dbl>, wk30 <dbl>,
# wk31 <dbl>, wk32 <dbl>, wk33 <dbl>, wk34 <dbl>, wk35 <dbl>, wk36 <dbl>,
# wk37 <dbl>, wk38 <dbl>, wk39 <dbl>, wk40 <dbl>, wk41 <dbl>, wk42 <dbl>,
# wk43 <dbl>, wk44 <dbl>, wk45 <dbl>, wk46 <dbl>, wk47 <dbl>, wk48 <dbl>, …- In this dataset, each observation is a song.
- The first three columns (
artist,trackanddate.entered) are variables that describe the song. - Then we have 76 columns (
wk1-wk76) that describe the rank of the song in each week.
Your turn #7: Pivoting
Use
pivot_longer()to tidy the data (Tip: Create the new variablesweekandrank). Assign the resulting data frame to a new data frame calledtidy_billboard.Use the new
tidy_billboarddata frame to calculate which song has been the longest on rank 1 (Tip: usefilter(),group_by()andsummarize())
05:00
Your turn #7: Pivoting
- Use
pivot_longer()to tidy the data (Tip: Create the new variablesweekandrank). Assign the resulting data frame to a new data frame calledtidy_billboard.
- Use the new
tidy_billboarddata frame to calculate which song has been the longest on rank 1 (Tip: usefilter(),group_by()andsummarize())